Kevin Rychel
I am a PhD bioengineer and data scientist specializing in ‘omics data analysis and computational biology. I have a strong interest in public health, synthetic biology, and drug discovery. I dream of making the world healthier and more sustainable.
About Me
During my PhD at UCSD (Spring 2023), I helped to lead a team that analyzed tens of thousands of samples of microbial RNAseq data. Using an unsupervised machine learning algorithm, we were able to group genes into independently modulated sets (iModulons), which reflect the biology of gene expression regulation and can be used to make discoveries and simplify data interpretation. We developed iModulon structures for over a dozen diverse species, several of which can be viewed via a web tool I built at ![]() iModulonDB.org. By interpreting gene expression data with this new tool, and combining it with other computational methods, I was able to reveal several systems-level mechanisms of stress tolerance in laboratory evolution studies. My thesis won the Engelson PhD Thesis Award 2023, which is given to the best overall PhD thesis from the UCSD Bioengineering department. Read my graduation story here!
iModulonDB.org. By interpreting gene expression data with this new tool, and combining it with other computational methods, I was able to reveal several systems-level mechanisms of stress tolerance in laboratory evolution studies. My thesis won the Engelson PhD Thesis Award 2023, which is given to the best overall PhD thesis from the UCSD Bioengineering department. Read my graduation story here!
I am very proud of my leadership experience and collaborations. In addition to my thesis work, I lead the "Genome Analytics" subgroup of my lab (Systems Biology Research Group, Bernhard Palsson), which included ~17 researchers. In this role, I mentored several graduate student projects, edited manuscripts, assisted with grant applications, liaised with collaborators, performed organizational tasks, and gave presentations. I worked closely with the Center for Biosustainability at DTU in Denmark, where the iModulon tools are applied for improving biomanufacturing applications.
I am an expert on gleaning useful biological knowledge and mechanistic predictions from large datasets and computational models. I am now applying these skills to track infectious diseases at Palmona Pathogenomics, where I build pipelines for training and deploying machine learning models and help to design a Bio-IT platform.
Who I Am in Three Words
Curious
I am deeply invested in understanding the world around me, including areas in and far outside of my expertise.
- Open-minded
- Creative
- Motivated by interest & passion
Charismatic
I love to connect with my teams and collaborators, as well as present data, teach, and mentor.
- Excellent communication skills
- Leadership experience
- Fun-loving & balanced
Kind
I aim to make the world better, at all scales – from my small daily interactions to my lifelong career aspirations.
- Friendly & altruistic
- Value diversity, equity, and inclusion
- Motivated to create a positive impact
My PhD & Publications
Below is a brief overview of my PhD dissertation. Each article marked with the icon represents a manuscript for which I am an author (according to the legend below). Click on the icons to access the articles. Last Updated: September 2023.
You will also notice video links ( ). All videos, including my entire PhD defense, are available in a later section.
|
First author, published (3)
|
First author, not yet peer-reviewed (1)
|
|
Contributing author, published (13)
|
Contributing author, not yet peer-reviewed (2+)
|
Aim 1: iModulons of Bacillus
- Showed that iModulons successfully summarize gene expression in a gram-negative, bioindustry-relevant organism
- Revealed 5+ novel hypotheses about sensory and regulatory systems in Bacillus subtilis
- Characterized global reallocation during sporulation
- A collaborator performed a metabolic modeling analysis (Tibocha-Bonilla et al.) that validated some of my hypotheses
Aim 2a: Build iModulonDB
- Implemented and deployed
 iModulonDB.org, a knowledgebase of microbial transcriptional regulation
iModulonDB.org, a knowledgebase of microbial transcriptional regulation - Demonstrated web data visualization expertise
- Furthered FAIR principles: our research is now findable, accessible, interoperable, and reusable
- Also helped develop another database, proChIPdb (Decker et al.)
Aim 2b: Expand iModulonDB
- Led Genome Analytics team, which analyzed thousands of RNAseq datasets to produce iModulons for 11+ species and a proteome (Patel et al.).
- Contributed to other detailed iModulon analyses, like a look at the tradeoff between stress and growth (Dalldorf et al.).
- Recruited 10 graduate students and mentored some student projects
- Built dashboards, consulted on data interpretation and presentation, and edited manuscripts for each project:
Computational Pipeline
Sastry et al., 2021
Sulfolobus acidocaldarius
Chauhan et al., 2021
Mycobacterium tuberculosis
Yoo et al., 2022
Psuedomonas aeruginosa (1)
Rajput et al., 2022a
Pseudomonas putida
Lim et al., 2022
Pseudomonas aeruginosa (2)
Rajput et al., 2022b
Salmonella enterica
Yuan et al., 2022
Escherichia coli
Lamoureux et al., 2023
Streptococcus pyogenes
Hirose et al., 2023
Aim 3a: Evolution of Reactive Oxygen Species Tolerance
- Build on prior work to establish & share iModulons by using them to gain novel insights on how genomes evolve
- Presented 6 key stress tolerance strategies from E. coli cells evolved to tolerate the oxidizing herbicide paraquat
- Enhanced interoperability between data types by uncovering relationships between mutations, iModulons, metabolism, and phenotypes
- Utilized an oxidative stress metabolic and expression flux balance model
- Developed skills and knowledge relevant to troubleshooting and engineering bioproduction strains
Aim 3b: Evolution of High Temperature Tolerance
- Discovered key stress tolerance strategies from E. coli cells evolved to tolerate high temperatures
- Analyzed transcriptomic responses to understand hypermutator strains
- Revealed changes to regulation which can only be understood in the context of large scale datasets
- Identified a putative function for uncharacterized genes yjfIJKL based on a variety of evidence including AlphaGo structure predictions
Education & Other Experience
PhD in Bioengineering (Sep 2018 - Apr 2023)
University of California San Diego
- Received Engelson PhD Thesis Award 2023 for best overall thesis from the Bioengineering department, according to criteria of originality, depth of analysis, and potential impact.
- Graduate student researcher & group leader in Dr. Bernhard Palsson's Systems Biology Research Group (see above).
- Teaching assistant for "Systems Biology: Analysis of Large Scale Biological Data" for two years. Check out the great feedback I received in 2021 and 2020!
- Finalist in Grad SLAM, a UC-wide challenge to explain complex research to non-technical audiences in 3 minutes.
- Prior to joining the SBRG, I completed a 3 month rotation in Dr. Stephanie Fraley's lab and co-authored a review article on cellular interactions with biomaterials (Scott et al.).
- GPA: 3.97/4.00
Bachelors of Science in Biomedical Engineering (Aug 2014 - Apr 2018)
University of Michigan Ann Arbor
- Majored in Biomedical Engineering, minored in Computer Science. GPA: 3.98/4.00
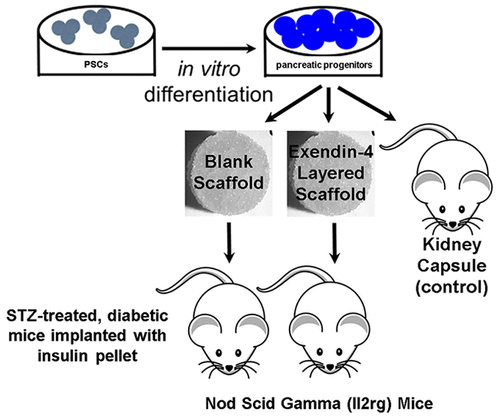
- Learned valuable skills during three years of research on a tissue engineered stem cell treatment for diabetes in Dr. Lonnie Shea's lab.
- Developed an image analysis pipeline and user interface which was used in a publication I co-authored (Kasputis et al.).
- Mentored a successful high school student's research project.
- Involved in outreach and volunteering through Tau Beta Pi.
Videos
I recommend using the full screen icon in the lower right corner of the video player.
PhD Defense [1:10:34]
I successfully defended my PhD at UCSD on April 24, 2023. In the video, I walk the audience through the general background, all three aims, and the conclusions of my work.
Grad Slam Finals [5:05]
I explain adaptive laboratory evolution and aim 3a for general audiences in 3 minutes with 3 slides. I won a $500 prize for this talk!
Genome Analytics [17:32]
I describe the Genome Analytics subgroup by walking through each of our databases. I explain iModulons breifly using examples from Bacillus (aim 1), then focus in on iModulonDB (aim 2) and ALEdb (aim 3). For audiences with some basic knowledge of biology.
Bacillus iModulons [6:44]
I walk you through my aim 1, for somewhat technical audiences.
Evolution Lecture [37:47]
I explain an evolution project in detail (aim 3a). This is a guest lecture in the Systems Biology course I used to TA, so this talk is for graduate students. The first half of this lecture may also be interesting to you if you just want to learn more about the iModulons of E. coli.
Skills, Techniques & Software
General Skills
- Data Analysis & Modeling
- Writing & Editing
- Presentations & Teaching
- Group Leadership
- Interactive Data Visualization
- Experimental Design
Programming
- Python
-
Pandas & NumPy
-
 Matplotlib & Seaborn
Matplotlib & Seaborn
-
Scikit-learn & SciPy
-
 Web Development
Web Development
-
 Bootstrap
Bootstrap
-
Highcarts & Tabulator
Data Types & Algorithms
- Transcriptomics
- Independent Component Analysis
- Genetics & Mutations
- Exometabolomic Rates
- Flux Balance Analysis
Biology Topics
- Transcriptional Regulation
- Stress Responses
- Sporulation
- Redox Metabolism
- Temperature Responses
- Cross-species Comparisons
- Genome Design
Some Experience & Ability to Learn
- Structural Analysis
- Databases & Cloud Computing
- Computational Pipelines
- Epigenetics
- Deep Learning
- Pangenomics & Genome Mining
Personal Life
I was originally inspired to become a bioengineer by my Mom, Lora. Her experience suffering from multiple sclerosis led to a dream to treat disease and improve lives. As I got older, and through interactions with the Center for Biosustainability in Denmark, I also became very interested in ways of using bioengineering to create sustainable solutions and fight climate change.
I live in San Diego with my incredible fiance, Sean. We love to visit the beach, hike, and travel as much as we can. I'm a proud member of the LGBT+ community.
Some of my hobbies include yoga, running, cooking, and digital art. I also do my best to keep up with all kinds of science news, and would always love to chat about space telescopes or AI!
I wrote this website from scratch using Bootstrap 5 and GitHub Pages. You can see the source code here. Some images were generated with Biorender, and many icons are from Font Awesome.
 About Me
About Me
 LinkedIn
LinkedIn
 Google Scholar
Google Scholar
 GitHub
GitHub
 Email
Email